Mmrf march 07 book.qxd
MMRF March 07 Book.qxd 4/16/07 8:33 AM Page 1 Clinical Medicine & ResearchVolume 5, Number 1: 1-7
2007 Marshfield Clinic
http://www.clinmedres.org
Original Research Use of an Electronic Medical Record
for the Identification of Research
Subjects with Diabetes Mellitus
Russell A. Wilke, MD, PhD; Richard L. Berg, MS; Peggy Peissig, MBA; Terrie Kitchner; Bozana Sijercic, MD; Catherine A. McCarty, PhD; and Daniel J. McCarty, PhD Diabetes mellitus is a rapidly increasing and costly public health problem. Large studies are needed tounderstand the complex gene-environment interactions that lead to diabetes and its complications.
The Marshfield Clinic Personalized Medicine Research Project (PMRP) represents one of the largestpopulation-based DNA biobanks in the United States. As part of an effort to begin phenotypingcommon diseases within the PMRP, we now report on the construction of a diabetes case-findingalgorithm using electronic medical record data from adult subjects aged ≥50 years living in one of thetarget PMRP ZIP codes. Based upon diabetic diagnostic codes alone, we observed a false positive caserate ranging from 3.0% (in subjects with the highest glycosylated hemoglobin values) to 44.4% (insubjects with the lowest glycosylated hemoglobin values). We therefore developed an improved casefinding algorithm that utilizes diabetic diagnostic codes in combination with clinical laboratory data andmedication history. This algorithm yielded an estimated prevalence of 24.2% for diabetes mellitus inadult subjects aged ≥50 years.
Keywords: Metformin; Natural language processing; Pharmacogenetics; Sulfonylurea
PMRP Working Group was formed to select diseases for whichelectronic algorithms could be developed to classify exposure and he current obesity epidemic represents a major outcome status using the electronic medical records contained international health problem.1 Genetic markers may be the most within the database. The diseases represent a range of anticipated efficient way to identify individuals at risk for obesity-related difficulty in using purely electronic methods to identify disease medical complications. One of the most costly obesity-related onset, disease progression and outcome. The first three diseases co-morbidities is diabetes mellitus (DM).2 Hyperglycemia is the were selected from a list of diseases that are routinely screened for clinical hallmark of DM, but the etiology of this heterogeneousdisorder likely involves multiple genetic and environmental during routine health maintenance examinations in adults. Listed interactions that ultimately result in alterations in insulin in order from expected greatest difficulty to least difficulty for secretion, insulin action or both.3,4 Large population-based electronic algorithms, the three diseases are: (1) glaucoma, (2) cohorts will be needed to characterize the genetics of complex osteoporosis, and (3) DM. The purpose of the current study was diseases such as DM.5,6 to pilot the process of electronically and manually abstractinginformation from the electronic medical record of adults served The Marshfield Clinic Personalized Medicine Research Project by Marshfield Clinic to define DM specifically, so that the PMRP (PMRP) is a population-based DNA biobank developed to database could eventually be utilized for studies designed to facilitate research in pharmacogenetics, genetic epidemiology characterize the genetic epidemiology and pharmacogenetics of and population genetics (www.mfldclin.edu/pmrp).7 In 2003, this disease.
the PMRP was mentioned in an article by Dr. Francis Collinsand colleagues from the National Human Genome Research Institute as it relates to their identified grand challenge to The current study protocol was approved by the Marshfield "develop robust strategies for identifying the genetic Clinic Institutional Review Board. The setting was a large contributions to disease and drug response."8 Therefore, a multi-specialty group practice located in central Wisconsin.
Reprint Requests: Russell A. Wilke, MD, PhD, Center for Human Genetics,
Received: October 10, 2006 Grant Support: This project was
Marshfield Clinic Research Foundation, 1000 North Oak Avenue, Revised: December 22, 2006 funded by a grant from Marshfield Marshfield, WI 54449, Tel: 715-389-3885, Fax: 715-389-3808, Accepted: January 8, 2007 Clinic Research Foundation.
MMRF March 07 Book.qxd 4/16/07 8:33 AM Page 2 The target population included residents within a single ZIP laboratory and radiology results. Since nearly everyone code (54449), encompassing the city of Marshfield (population residing in the target ZIP code for the current study receives 19,000). This ZIP code was selected because nearly everyone their health care through Marshfield Clinic, this record is in the population seeks their health care through Marshfield Clinic, a fully integrated health care system with a long-standing comprehensive electronic medical record.9 The target Study Population ZIP code was also one of 19 ZIP codes selected to recruit Subjects were considered eligible for this study based on the subjects for the Marshfield Clinic PMRP.7 following criteria: (1) age 50 years or older, (2) alive onDecember 31, 2002, (3) seen at Marshfield Clinic between Briefly, PMRP is a large biobank containing DNA and sera January 1, 2000 and December 31, 2002, and (4) residing in ZIP from approximately 19,000 Marshfield Clinic patients. Each code 54449 (Marshfield). Electronic medical record data for the PMRP participant has also provided informed consent eligible subjects were searched to determine the presence (or allowing their genetic and serologic data to be linked to all absence) of diabetes diagnostic codes from the International available clinical data within their electronic medical record Classification of Diseases, Ninth Revision (ICD-9 codes). These using a confidential and secure encryption process. PMRP codes included primary diagnostic codes for diabetes (ICD-9 therefore provides a unique opportunity to conduct very large codes 250.00-250.92), and secondary diagnostic codes for genetic studies on a variety of common diseases.
diabetic neuropathy (ICD-9 code 357.2), retinopathy (ICD-9codes 362.01-362.02) and nephropathy (ICD-9 code 583.81).
Medical Record For each potential study subject, clinical laboratory data were Electronic medical records have been utilized at Marshfield scanned electronically to identify relevant test results. These Clinic since the 1960s, and the vast majority of patient included all available glucose and glycosylated hemoglobin records within this system have been electronic for over a (HbA1c) values. Each glucose value was assumed to be random decade. A variety of data are captured. One of the key features (i.e., non-fasting) unless otherwise specified. Maximum values of the Marshfield Clinic electronic medical record is a were determined for each subject.
Windows application called the combined medical record(CMR). CMR integrates data from all Marshfield Clinic facilities and cooperating hospitals, including Saint Joseph's We have previously utilized natural language processing Hospital (Marshfield). CMR includes indices to all events (NLP) software to reconstruct complete retrospective and encounters that patients have experienced within the medication use histories for all research subjects participating Marshfield Clinic system of care, and it can be used to access in the PMRP Biobank.10 We have also shown previously that all textual documentation such as office notes, operative these data are amenable to electronic abstraction, and that reports and discharge summaries. CMR also includes they can be managed programmatically to yield high quality comprehensive lists of patient problems, a summary of each drug exposure histories in the context of lipid lowering clinic encounter (diagnoses and procedures), a variety of therapy (e.g., 100% sensitive and 96% specific, with a medication alerts, and online access to over a decade of precision of 95%).11 In the current study, clinic records from Table 1. Electronically abstracted text mention of glucose lowering medication* for the entire study cohort (n=8101).
Diagnostic Code Available: Laboratory Data Available: * Drug code: I, insulin; M, metformin; S, sulfonylurea.
Diabetes case f inding CM&R 2007 : 1 (March)
MMRF March 07 Book.qxd 4/16/07 8:33 AM Page 3
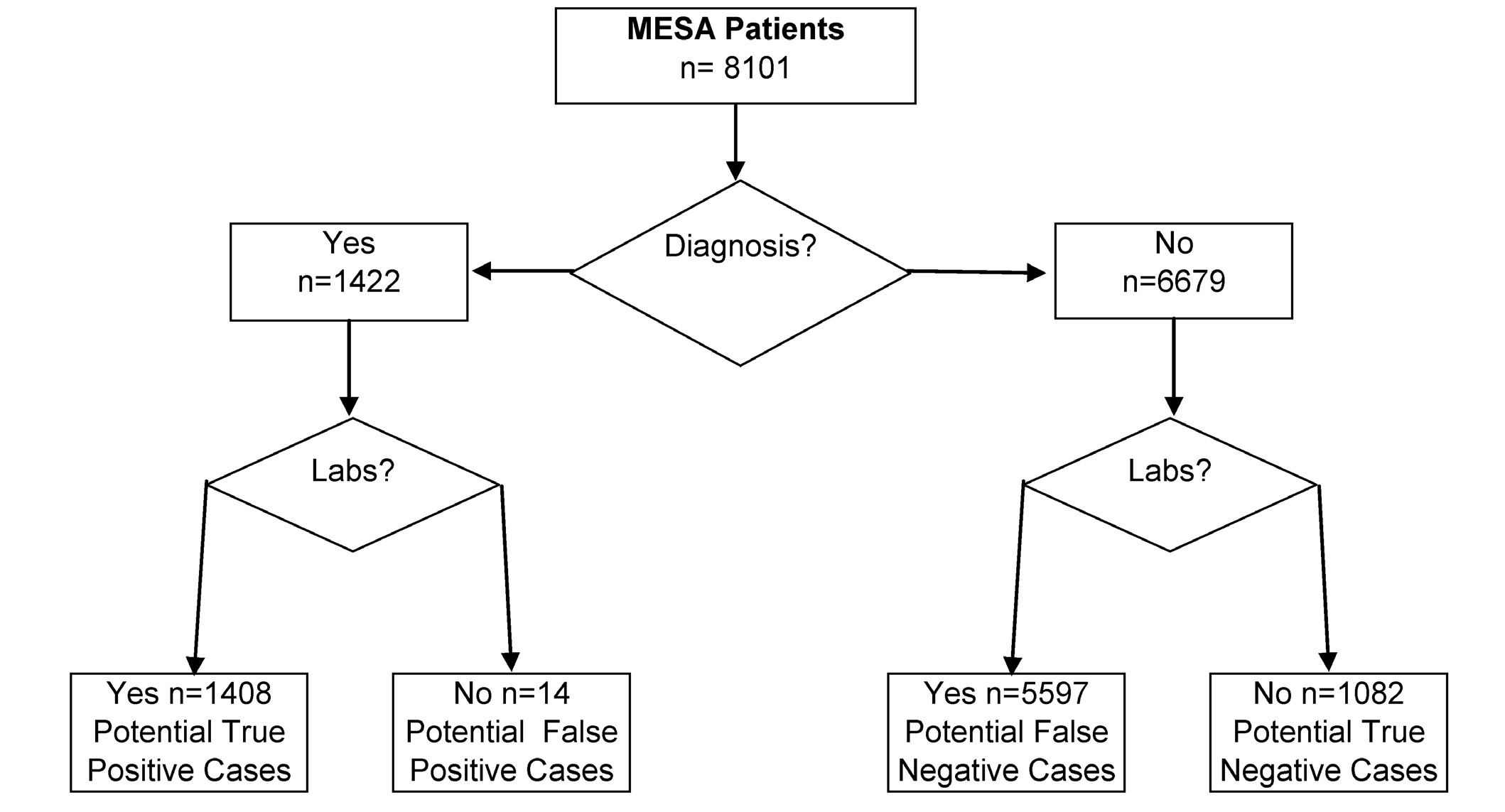
Figure 1. Initial electronic classification of study subjects based upon two criteria: first, the presence or absence of diabetic
diagnostic codes, and second, the presence or absence of relevant clinical laboratory data (e.g., glucose levels and glycosylated
hemoglobin [HbA1c] levels). This strategy produced four data bins. These bins contain 1408, 14, 5597 and 1082 study subjects,
respectively (total study population 8101). MESA = Marshfield Epidemiologic Study Area.
all eligible subjects were re-interrogated electronically for
specific subjects with records containing diabetic diagnostic
text mention of three classes of glucose lowering medications.
codes but no corresponding laboratory data. Research
This involved the application of NLP software entitled
coordinators were also asked to manually abstract data from a
FreePharma (Language & Computing; http://www.landc.be).
specific sample of subjects without electronic diabetic
All 8101 subject records were searched electronically to
diagnostic codes: 72 subjects who had the most extreme
identify and catalogue dates for all text notes mentioning any
glucose or HbA1c results. American Diabetes Association
sulfonylurea agent known to be commercially available
(ADA) diagnostic criteria were used to confirm the presence
within the past decade. This included four "first-generation"
or absence of DM (i.e., fasting glucose ≥126 on two
sulfonylureas (acetohexamide, chlorpropamide, tolbutamide,
occasions or a single random glucose >200).
tolazamide) and three "second-generation" sulfonylureas(glimepiride, glipizide, glyburide). A similar approach was
taken to identify all text notes containing mention of any
The study population included 8101 patients who met the
therapeutic agent mapping to the generic drug names
inclusion criteria. This number is comparable to the year 2000
metformin (the only clinically approved glucose-lowering
US Census estimate (n = 7905) for this age group and ZIP
biguanide) and insulin (table 1).
code. All medical records from these study subjects wereinterrogated electronically for the presence of diagnostic
codes associated with DM. Of the 8101 study subjects, 1422
A five-page data abstraction form was developed for use by
(17.6%) subjects were found to have at least one diabetic
trained research coordinators to manually abstract data
diagnostic entry, i.e., either diabetes or a diabetic end organ
related to DM diagnosis and treatment from the medical
complication (figure 1, left). The remaining medical records
records. This form was used to collect demographic data and
(n = 6679 study subjects) had no diabetic diagnostic entries
specific diabetes-related clinical information. For quality
(figure 1, right). Each of these two initial subsets (1422 with
assurance, 10% of all manually abstracted records were re-
codes and 6679 without) is discussed separately below in the
abstracted by a second research coordinator, and
context of phenotyping accuracy.
discrepancies resolved by a licensed practicing physician.
Research coordinators were asked to manually abstract data
Diagnostic Codes Present
for three sets of subjects with electronically recognized
Among the 1422 study subjects with at least one diabetic
diabetic diagnostic codes: (1) 100 subjects with the highest
diagnostic code, 99% (1408 study subjects) had sufficient
HbA1c, (2) 100 subjects with the lowest HbA1c, and (3) 14
clinical laboratory data to either support or refute the
CM&R 2007 : 1 (March)
Wilke et al.
MMRF March 07 Book.qxd 4/16/07 8:33 AM Page 4
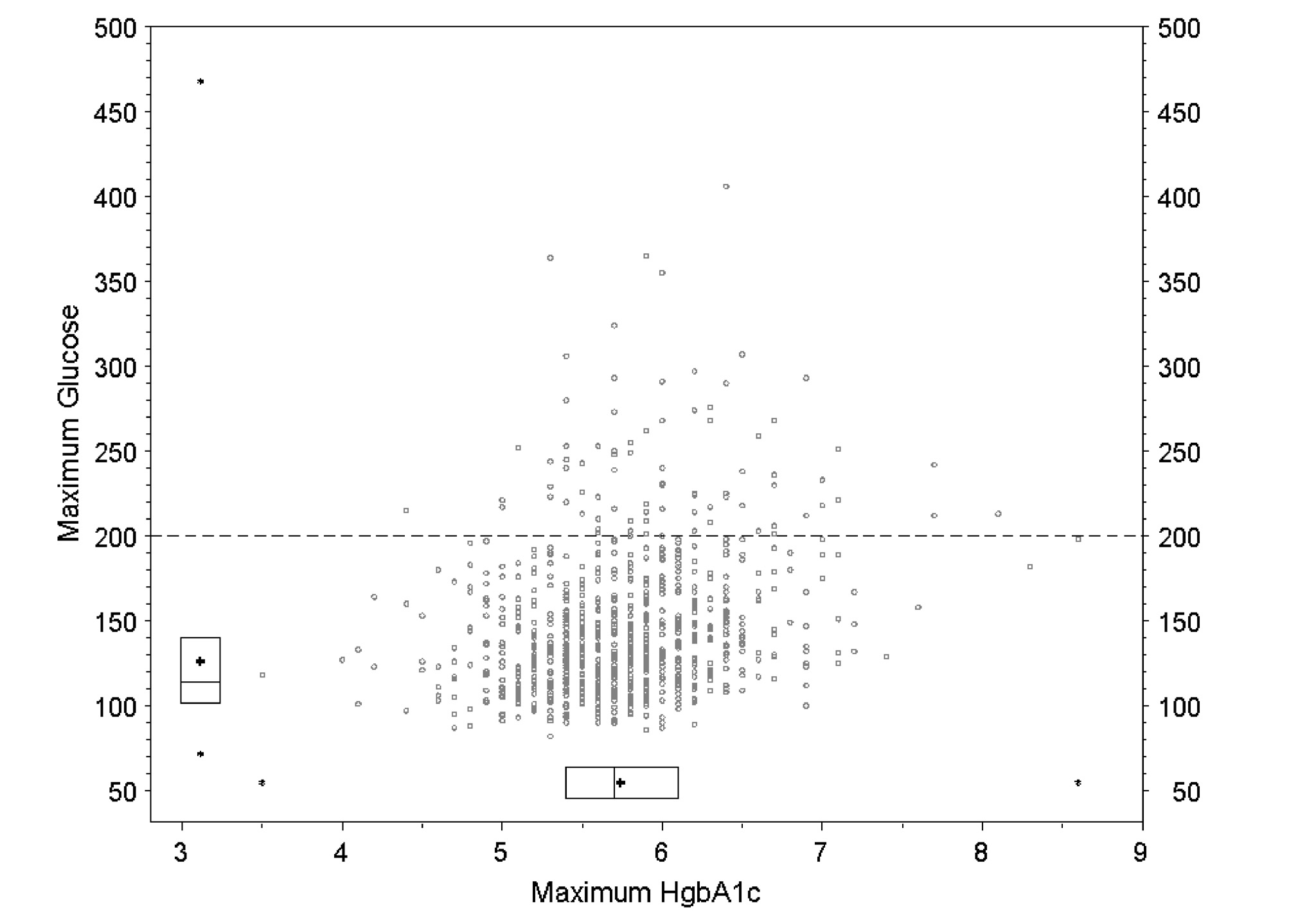
Figure 2. Graphic representation of the clinical laboratory data from bin 3 (n = 5597), as classified in the text (potential false
negative cases) and illustrated in figure 1. Glucose levels and glycosylated hemoglobin (HbA1c) levels were abstracted
electronically for all 5597 subjects in bin 3. The 854 patients who had at least one glucose level and at least one HbA1c level
are shown as circles in the scatter plot. The box plots in the margins reflect all available data. The mean is shown as a "+" within
boxes representing the 25th, 50th and 75th percentiles. Asterisks denote minimum and maximum. The dashed horizontal line
indicates a glucose level 200 mg/dl.
diagnosis. These data include fasting glucose, random
at least one glucose value or at least one HbA1c level. Since
glucose or HbA1c levels. The1408 subject records containing
it was likely that some of these 5597 potential false negative
these data represent potential true positive cases of DM
cases were actually either undiagnosed diabetics or diabetics
(figure 1). Based upon manual data abstraction, we observed
treated without a corresponding provider-entered diagnostic
that diagnostic codes yielded a true positive rate for DM
code, relevant clinical laboratory data were re-abstracted
ranging from 97.0% (in 100 subjects with the highest HbA1c
electronically for all 5597 subjects. These clinical laboratory
values) to 55.6% (in 100 subjects with the lowest HbA1c
data are summarized in figure 2. For both axes (glucose and
values). It should also be noted that our initial electronic
HbA1c), the mean is represented by a "+" located within box
screening strategy (e.g., diagnostic codes and laboratory
plots corresponding to the 25th, 50th and 75th percentiles,
data), as shown in figure 1, also yielded 14 potential false
respectively, for the entire dataset (n = 5597). The horizontal
positive cases of DM (i.e., diabetic diagnostic codes without
dashed line delineates a glucose level ≥200 mg/dl.
any supporting electronic laboratory data). Among these 14potential false positive cases of DM, four were manually
Within figure 2, only those 854 subjects found to have both a
confirmed to be diabetic based upon treatment history or
glucose level and an HbA1c level have been represented as
laboratory data not available electronically.
circles in the scatter plot. Data were manually abstracted for72 study subjects with the most extreme glucose and HbA1c
Diagnostic Codes Absent
values. Of these, 41 records contained a glucose value >200
Electronic interrogation of the entire medical record for each
mg/dl. All 41 records (100%) were manually confirmed as
of the 8101 unique subjects in this study revealed that 6679 of
cases of DM.
these subjects had no diabetic diagnostic codes containedwithin their electronic medical record (figure 1, right side).
Of the 8101 unique subjects in this study with no diabetic
Of these, 5597 (84%) had clinical laboratory data containing
diagnostic codes (figure 1, right side), 1082 (16%) had no
Diabetes case f inding
CM&R 2007 : 1 (March)
MMRF March 07 Book.qxd 4/16/07 8:33 AM Page 5
clinical laboratory data that could be used to discriminate
Present versus Diagnostic Codes Absent) suggest that the first
between diabetic and non-diabetic (i.e., no glucose levels and
branch point in this algorithm can be based upon diagnostic
no HbA1c levels). These 1082 subjects are assumed to be true
codes. The two subsequent branches of the algorithm then
negative cases (i.e., not diabetic). The design of this study
apply differential logic, reflecting the following two
(retrospective chart review) does not allow the discrimination
assumptions. First, in the situation where diabetic diagnostic
of false negative cases within this specific sub-sample
codes are present, any purely electronic algorithm simply needs
because the research subjects were neither interviewed nor
to confirm the diagnosis. This can be done by documenting
examined during the conduct of the study. However, this
either abnormal laboratory data (HbA1c>ULN, or glucose
population is known to be highly compliant with primary
criteria established by the ADA) or treatment with one of three
prevention screening visits.12 Among the 5597 potential false
known medications used as first line therapy for DM.
negative case subjects with laboratory data but no diagnostic
Conversely, in the situation where diabetic diagnostic codes are
codes, 4477 (80%) were found to have at least one glucose
absent, the algorithm needs to establish the diagnosis. Since
level within 2 years. Based upon these observations, and the
this latter step is more than simply confirmatory, the rightward
additional observation that patients residing in the target
arm of the algorithm needs to be sufficiently stringent to
study ZIP code receive nearly all their healthcare (90% of
minimize (and, if possible, avoid altogether) false positive
outpatient visits, 95% of inpatient visits) through Marshfield
case assignment. Based upon the distribution of laboratory
Clinic,9 it is reasonable to assume that the frequency of false
data observed in figure 2 (sub-sample with n=5597), we
negative cases would be low in the sub-sample of 1082
recommend that the identification of false positive case
subjects with no relevant clinical laboratory data.
subjects within this sub-sample be made by first using thepresence of an HbA1c test to suggest a reasonable clinical
index of suspicion for DM, and then, second by accepting a
We propose the electronic case-finding algorithm shown in
maximum glucose value >200 mg/dl as diagnostic.
figure 3. The observations outlined above (Diagnostic Codes
Figure 3. Proposed algorithm for identification of case subjects with diabetes mellitus (DM) in the Personalized Medicine
Research Project (PMRP) database.
CM&R 2007 : 1 (March)
Wilke et al.
MMRF March 07 Book.qxd 4/16/07 8:33 AM Page 6
The final electronic algorithm was used to identify unique
patients with DM. This electronic algorithm was applied to
The current study presents an electronic case-finding
the entire study cohort (8101 adults aged ≥50 years and living
algorithm that can be used for the identification of research
in the target ZIP code), identifying 1960 (24.2%) unique
subjects with DM in the PMRP DNA biobank. It is important
subjects with DM.
to note that DM is a clinically heterogeneous disorder, andthat the current study does not discriminate between major
forms of the disease. No effort was made to sub-classify
The current study outlines the construction of a case finding
subjects identified by this algorithm according to major type
algorithm for DM. This algorithm utilizes diagnostic codes,
(e.g., type 1 versus type 2 diabetes), or minor type (e.g.,
clinical laboratory data and medication history to identify
maturity onset diabetes of the young or latent autoimmune
subjects with DM in a large patient database. Using
diabetes in adults). It is anticipated that further phenotypic
diagnostic codes alone, we observed a high rate of false
discrimination will be accomplished, on a study-by-study
positive cases. Further confirmation is therefore required
basis, during future applications of this algorithm using
through clinical laboratory data (using current ADA
context-specific parameters defined by each study.
diagnostic criteria3) or medication data (obtained by NLP10).
By considering these additional data, the final algorithm
reduces the frequency of false positive cases.
The authors thank Ms. Laura Lobner for data collection andquality assurance, Ms. Carla Rottscheit for data management
The final algorithm also reduces the frequency of false negative
and Ms. Ekta Sirohi for the electronic abstraction of
cases by identifying subjects with DM in the absence of a
medications. We also thank Marshfield Clinic Research
diabetic diagnostic code. However, this portion of the algorithm
Foundation for its support through the assistance of Alice
is conservative in that it requires the presence of an elevated
Stargardt and Linda Weis in the preparation of this
random glucose level (≥200 mg/dl) specifically within the
context of a subject record also containing at least one HbA1cvalue. We opted not to accept an elevated glucose level alone,
The Working Group for the Personalized Medicine Research
since in the absence of diagnostic codes for diabetes, a random
Project Phenotyping Engine includes Dr. Philip Giampietro,
glucose value can be elevated for a variety of non-diagnostic
Dr. Robert Greenlee, Dr. Catherine McCarty, Dr. Daniel
reasons (e.g., steroid therapy or intravenous fluid replacement
McCarty, Ms. Peggy Peissig and Dr. Russell Wilke.
containing dextrose). Since the presence of at least one HbA1ctest (whether normal or elevated) indicates an increased clinical
index of suspicion for DM, an elevated random glucose level
1. Mokdad AH, Ford ES, Bowman BA, Dietz WH, Vinicor F, Bales VS,
can be considered diagnostic in this context. Although
Marks JS. Prevalence of obesity, diabetes, and obesity-relatedhealth risk factors, 2001. JAMA 2003; 289:76-79.
stringent, our inclusion of a strategy to reduce false negative
2. Fontaine KR, Bartlett SJ. Access and use of medical care among
cases was necessary in this study population because the
obese persons. Obes Res 2000; 8:403-406.
Centers for Disease Control and Prevention have estimated that
3. American Diabetes Association. Diagnosis and classification of
a significant proportion of all adult diabetic subjects in the
diabetes mellitus. Diabetes Care 2006; 29(suppl 1):S43-S48.
United States remain undiagnosed.13
4. Wilke RA, Jochen AL, Maas DL and IM O'Shaughnessy:
Hypoglycemia and Diabetes Mellitus. In: Kutty K, SebastianJL, Mewis BA, Berg DD, Kochar, MS, eds. Kochar's concise
Application of the final algorithm yielded an estimated DM
textbook of medicine. 3rd ed. Baltimore, MD: Williams &
prevalence of 24.2% for adults aged ≥50 years residing in the
Wilkins; 1998.
target ZIP code (i.e., the algorithm identified 1960 of the
5. Kaiser J. Biobanks. Population databases boom, from Iceland to
8101 study subjects as diabetic case subjects). The prevalence
the U.S. Science 2002; 298:1158-1161.
6. Davis RL, Khoury MJ. The journey to personalized medicine.
of DM is highly associated with age, and our observation is
Personalized Med 2005; 2:1-4.
consistent with previously published estimates.13-15 This
7. McCarty C, Wilke RA, Giampietro PF, Wesbrook SD, Caldwell
work adds to a growing body of literature supporting the
MD. Marshfield Clinic Personalized Medicine Research
utility of electronic medical records for case-finding
Project (PMRP): design, methods and recruitment for a large
specifically within the context of DM.16-18 Further, the
population-based biobank. Personalized Med 2005; 2:49-79.
8. Collins FS, Green ED, Guttmacher AE, Guyer MS; US National
current study extends these observations through the
Human Genome Research Institute. A vision for the future of
development of an electronic algorithm that considers clinical
genomics research. Nature 2003; 422:835-847.
laboratory data and medication history in addition to
9. Greenlee RT. Measuring disease frequency in the Marshfield
diagnostic codes. Since the target ZIP code characterized in
Epidemiologic Study Area (MESA). Clin Med Res 2003;
the current study is located within the geographic region
10. Sirohi E, Peissig P. Study of effect of drug lexicons on
represented by the Marshfield Clinic PMRP database, the
medication extraction from electronic medical records. Pac
resulting algorithm will be useful for identifying DM cases in
Symp Biocomput 2005; 10:308-318.
this database.
Diabetes case f inding
CM&R 2007 : 1 (March)
MMRF March 07 Book.qxd 4/16/07 8:33 AM Page 7
11. Peissig P, Sirohi E, Berg RL, Brown-Switzer C, Ghebranious N,
Catherine A. McCarty, PhD
McCarty CA, Wilke RA. Construction of atorvastatin
Center for Human Genetics
dose-response relationships using data from a large
Marshfield Clinic Research Foundation
population-based DNA biobank. Basic Clin PharmacolToxicol 2007; 100:286-288.
1000 North Oak Avenue
12. McCarty CA, Chyou PH, Greenlee R, McCarty DJ, Gunderson
Marshfield, Wisconsin 54449
P, Reding D. Differences in preventive screening rates inWisconsin farm and non-farm resident women. WMJ 2003;
Bozana Sijercic, MD
Department of Internal Medicine
13. National Diabetes Fact Sheet. Centers for Disease Control and
Prevention Web site. Available at:
Marshfield Clinic
1000 North Oak Avenue
Accessed September 21, 2005.
Marshfield, Wisconsin 54449
14. Flegal KM, Carroll MD, Ogden CL, Johnson CL. Prevalence
and trends in obesity among US adults, 1999-2000. JAMA
Daniel J. McCarty, PhD
15. Harris MI, Flegal KM, Cowie CC, Eberhardt MS, Goldstein
Marshfield Epidemiology Research Center
DE, Little RR, Wiedmeyer HM, Byrd-Holt DD. Prevalence of
Marshfield Clinic Research Foundation
diabetes, impaired fasting glucose, and impaired glucose
1000 North Oak Avenue
tolerance in U.S. adults. The Third National Health and
Marshfield, Wisconsin 54449
Nutrition Examination Survey, 1988-1994. Diabetes Care1998; 21:518-524.
16. Hassey A, Gerrett D, Wilson A. A survey of validity and utility
of electronic patient records in a general practice. BMJ 2001;322:1401-1405.
17. Szeto HC, Coleman RK, Gholami P, Hoffman BB, Goldstein
MK. Accuracy of computerized outpatient diagnoses in aVeterans Affairs general medicine clinic. Am J Manag Care2002; 8:37-43.
18. Newton KM, Wagner EH, Ramsey SD, McCulloch D, Evans R,
Sandhu N, Davis C. The use of automated data to identifycomplications and comorbidities of diabetes: a validationstudy. J Clin Epidemiol 1999; 52:199-207.
Author Affiliations
Russell A. Wilke, MD, PhD
Center for Human Genetics
Marshfield Clinic Research Foundation and
Department of Internal Medicine
Marshfield Clinic
1000 North Oak Avenue
Marshfield, Wisconsin 54449
Richard L. Berg, MSBiomedical Informatics Research CenterMarshfield Clinic Research Foundation1000 North Oak AvenueMarshfield, Wisconsin 54449
Peggy Peissig, MBABiomedical Informatics Research CenterMarshfield Clinic Research Foundation1000 North Oak AvenueMarshfield, Wisconsin 54449
Terrie KitchnerCenter for Human GeneticsMarshfield Clinic Research Foundation1000 North Oak AvenueMarshfield, Wisconsin 54449
CM&R 2007 : 1 (March)
Wilke et al.
Source: http://www.cs.uwm.edu/classes/cs870/article3.pdf
smar.ma
Pédiatrie 1 P1- Antibiothérapie probabiliste en milieu de réanimation pédiatrique O.EL ALLAM, Y.HARTI, Y.ALAOUI, B.HMAMOUCHI, S.NEJMI, A.CHLILEK SERVICE DE REANIMATION PEDIATRIQUE POLYVALENTE CHU IBN ROCHD DE CASABLANCA Introduction : L'antibiothérapie probabiliste correspond à une prescription d'antibiotiques réalisée avant de connaitre la nature et la sensibilité des germes responsable de l'infection. En pédiatrie l'évolution d'un processus infectieux sévère est souvent plus rapide que chez l'adulte, avec le risque d'apparition souvent précoce d'une insuffisance circulatoire. Le but de notre travail est la description et l'évaluation de l'antibiothérapie probabiliste en milieu de réanimation pédiatrique polyvalente CHU Ibn Rochd de Casablanca. Patients et méthodes : Etude rétrospective étalée sur 11 mois de janvier 2012 à novembre 2012 qui a permis le recrutement de 142 patients. Les données recueillies sont les critères épidémiologiques des patients, les antécédents médicaux, la notion de colonisation bactérienne, le type d'infection motivant l'introduction de l'antibiothérapie probabiliste, les circonstances du choix de l'antibiotique, le caractère précoce ou tardif et la durée de l'antibiothérapie probabiliste, le retentissement du changement de l'antibiothérapie sur le pronostic, l'évolution et la durée de séjour. Résultats : L'âge moyen était de 37,44 mois, le poids moyen était de 13,28kg, 7,7%des patients avaient des antécédents cardiaques, 4,2% avaient des antécédents respiratoires, 1,4% avaient un déficit immunitaire, 1,4% étaient anciens prématurés, 83,1% des patients étaient hospitalisés antérieurement avec notion de prise d'antibiotiques dans 11,3% des cas. 64,8% de nos patients avaient une infection pulmonaire, 9,2% avaient une infection urinaire, 13,4% avaient une infection neuromeningée, 15,5% une septicémie. L'antibiothérapie probabiliste prescrite était à base d'une monothérapie dans 11,5% des cas, une bithérapie dans 59,8% des cas et une trithérapie dans 28,7% des cas avec le choix du ceftriaxone dans 60,5% des cas. L'heure de début de l'antibiothérapie était le jour dans 52,8% des cas, la nuit dans 44,4% et le weekend dans 2,8% des cas. La décision était prise par un médecin junior dans 54,2% des cas et un médecin seigneur dans 45,8% des cas avec un changement de cette antibiothérapie selon la gravité dans 38% des cas et selon la bactériologie dans 21,8% des cas. La durée moyenne de l'antibiothérapie probabiliste était de 10,79 jours. L'évolution était favorable dans 66,2% des cas avec un taux de mortalité de 33,8%. Conclusion : La prescription raisonnée de l'antibiothérapie probabiliste initiale a démontré son impact sur l'amélioration du pronostic vital des patients. Le caractère nosocomial ou communautaire de l'infection, la connaissance de l'écologie bactérienne du service où l'on travaille, de la flore colonisante du patient et des données de l'examen direct des prélèvements bactériologiques jouent un rôle majeur dans cette décision.
math.helsinki.fi
Second order logic and set theory Both second order logic and set theory can be used as a foundation for mathematics, that is, as a formal language in which propositions ofmathematics can be expressed and proved. We take it upon ourselvesin this paper to compare the two approaches, second order logic on onehand and set theory on the other hand, evaluating their merits andweaknesses. We argue that we should think of first order set theoryas a very high order logic.